Transcription-Associated Cas9 Targeting in Eukaryotic Cells
A novel mechanism for conditional spatiotemporal control of Cas9 activity without modification of its core protein domains, gRNA components, or expression levels.
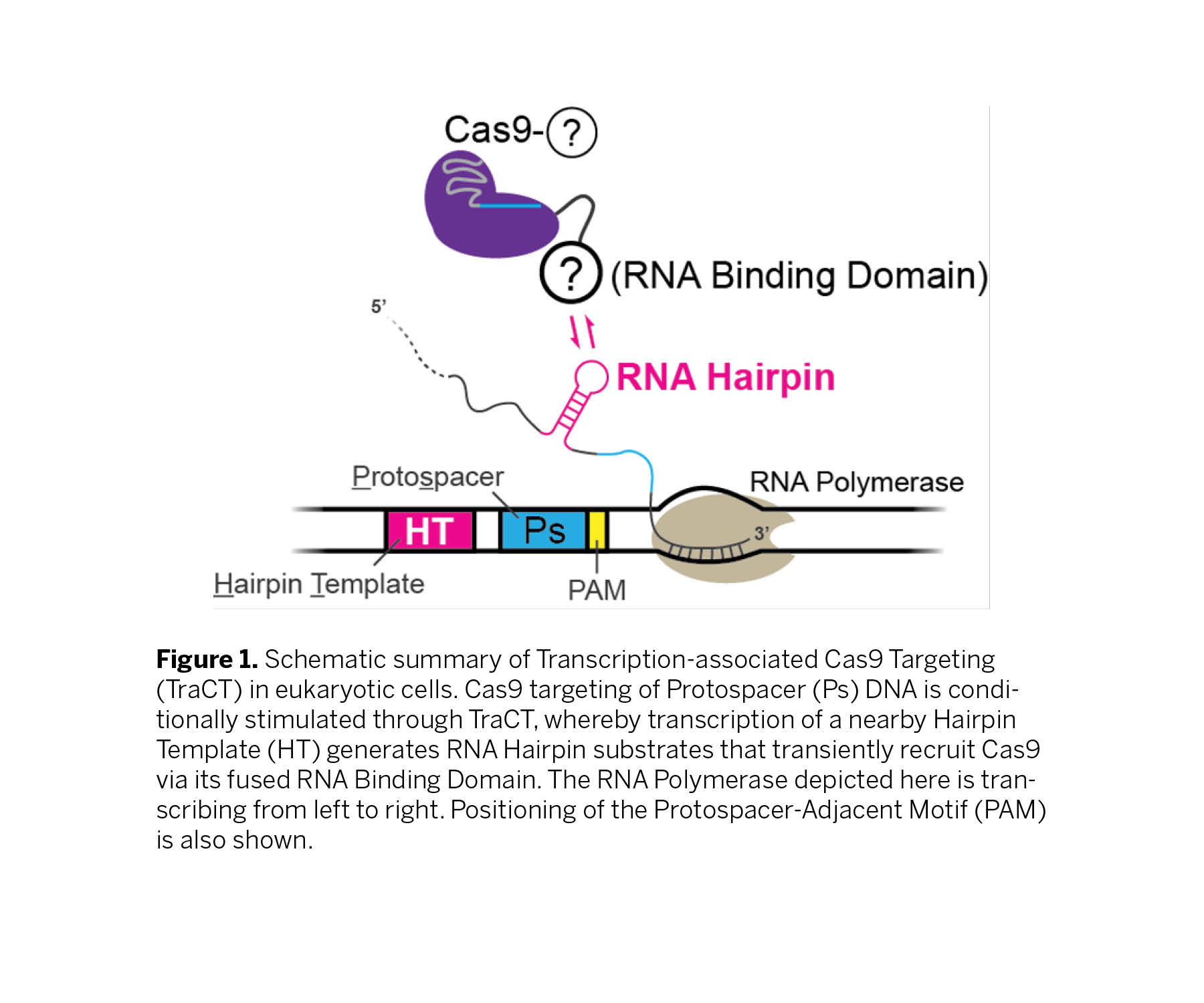
Technology
NYU Langone Health researchers have engineered novel Cas9 fusions that conditionally interact with hairpin template (HT) elements near target sites, enhancing targeting when suboptimal PAM interactions limit activity. Using a novel Transcription-associated Cas9 Targeting (TraCT) mechanism, they have demonstrated control over gene editing activity, influenced by the orientation of the protospacer (Figure 1). Furthermore, the level of HT transcription dynamically scaled TraCT activity, generating allelic editing preference for highly transcribed alleles. This implies that TraCT could be applied for cell type-specific editing in animal cell populations with differentially transcribed HTs. Importantly, the team demonstrated that TraCT can function in human cells, making it a potentially useful tool for human gene editing. The TraCT innovation also includes a generalizable yeast two-hybrid platform to test for co-transcriptional protein-RNA interactions that does not require labor-intensive hi-res live-cell imaging or immunoprecipitation procedures. In conclusion, this technology offers a dynamic and precise method of gene editing, with potential applications in gene editing and gene therapy. The experiments conducted have demonstrated its efficacy and versatility, making it a promising tool for future research and development.
Background
Class 2 CRISPR-associated protein, Cas9, is a gene-editing tool that has surged in popularity due to its ease and dynamic use in removing, adding and altering DNA sequences. Cas9 takes advantage of guide RNAs (gRNAs) to locate and cut DNA at desired locations. CRISPR-Cas sector market size was $3.1B in 2022 with a 5-year compound annual growth rate (CAGR) of 26.3%. As this tool gains popularity, its importance in selective and accurate gene editing becomes more prominent for utilizing this method in finding new therapies. This novel method provides a spatiotemporal control tool in gene editing approaches: transcription-associated Cas9 targeting in eukaryotic cells. The transcription-associated and spatiotemporal control of Cas9’s targeting ability can be achieved without modifying the protein’s core domains, gRNA components, or their expression levels.
Applications
- Therapeutic applications: The treatment of genetic diseases where precise timing and dosage of gene editing are crucial (e.g. Personalized medicines)
- Enables allelic editing in a diploid
- In cases where a mutation could result in the production of a harmful substance, it allows for the specific knockout of an allele
- It offers the ability to selectively edit one of two identical targets if the two alleles are transcribed at different levels
- Could be deployed to inactivate a highly expressed deleterious allele
Advantages
- Control over the spatial and temporal activity of Cas9 and conditional targeting can be achieved without necessitating any alterations to the protein’s central domains, the core gRNA components, or their levels of expression
- Increased gene editing precision means it could be applied in more complex biological cells (i.e., eukaryotes)
- Can be used for transcription-dependent allelic editing and cell type-specific editing in animal cell populations
- Widely applicable in different areas of biotechnology, such as genetic engineering and gene therapy
Intellectual Property
Provisional patent application pending
-
expand_more mode_edit Authors (3)Jef Boeke, PhDGregory Goldberg, PhDMarcus Noyes, PhD
-
expand_more cloud_download Supporting documents (2)Product brochureTranscription-Associated Cas9 Targeting in Eukaryotic Cells.pdfMarketing BriefNYU - Transcription-Associated Cas9 Targeting in Eukaryotic Cells - Marketing Brief - BOE01-20.pdf (83 KB)