KaryoCreate: Generating Chromosome-Specific Aneuploidies in Human Cells
Chromosome-specific aneuploidies on the cellular level.
Technology
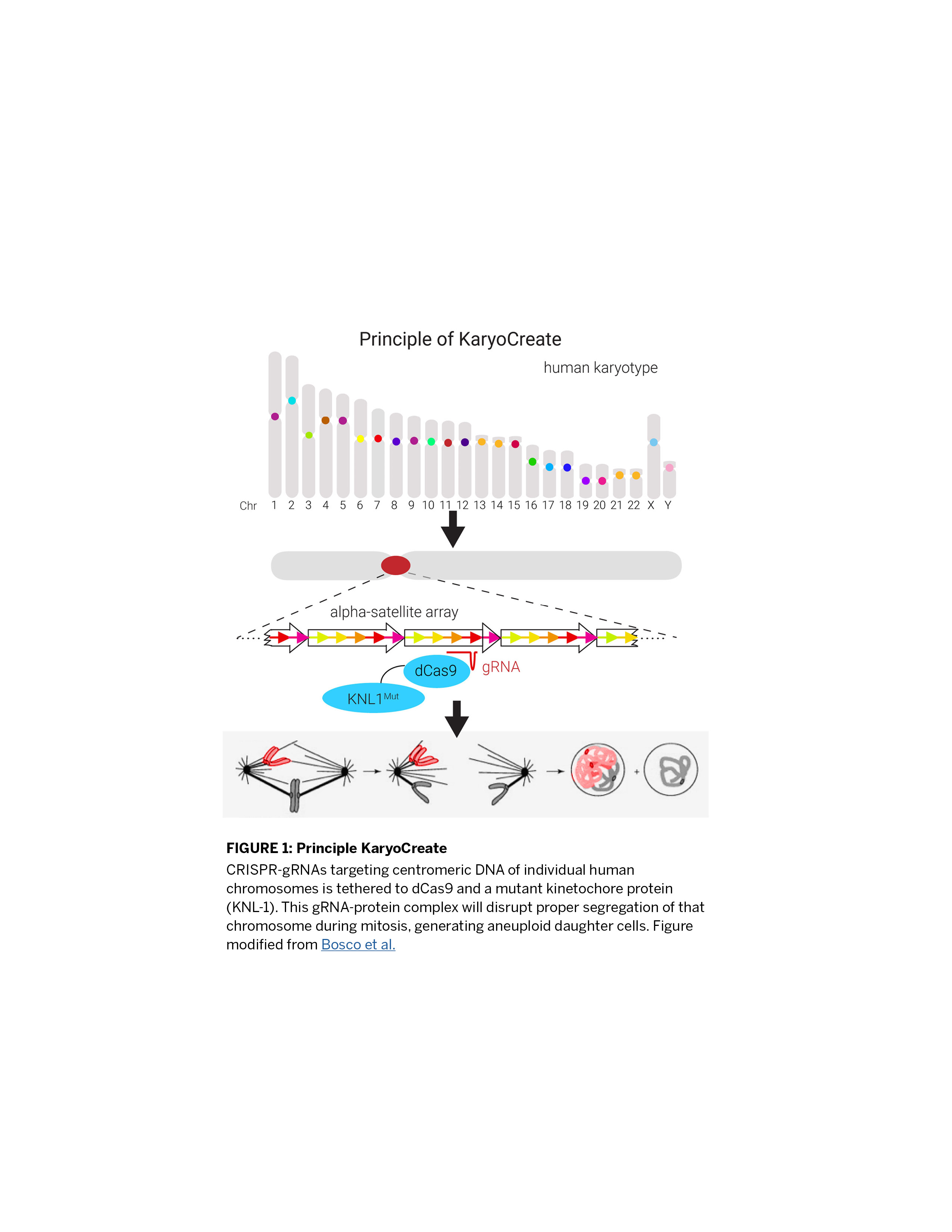
NYU’s KaryoCreate (Karyotype CRISPR-engineered aneuploidy technology) has revolutionized cell-based studies of aneuploidy. This system enables the creation of specific chromosome losses or gains in any cultured human cell line. With KaryoCreate, specific chromosome mis-segregations are generated via a CRISPR gRNA that targets chromosome-specific centromeric DNA sequences and is tethered to dCas9 and a mutant kinetochore protein (KNL-1) (Figure 1). The inventors used a proprietary algorithm to design gRNAs targeting centromeric DNA on 21 human chromosomes. They have validated 10 of these gRNAs in human cell lines, observing an average mis-segregation efficiency of approximately 8% for chromosome gains and 10% for chromosome losses. The inventors demonstrated the power of this system for studying cancer-specific aneuploidies by inducing chromosome 18q loss in colon cancer cells, which revealed that loss of this chromosome promotes resistance to TGF-β. These results can be found in their recent publication in Cell.
Background
Aneuploidy is defined as the presence of chromosome losses or gains and is a hallmark of cancer. Indeed, more than 90% of solid tumors contain chromosome number aberrations. Until now, studying aneuploidy has been challenging as there are few aneuploid cell models and existing methods for generating aneuploidies are inefficient and technically challenging. KaryoCreate technology vastly improves upon the efficiency and ease of existing techniques for studying aneuploidy with the additional benefit of targeting a specific chromosome for loss/gain. All human chromosomes contain a centromere, which is a long, repetitive region of DNA where cells assemble the kinetochore, a complex of proteins that regulate chromosome segregation. Specifically, each kinetochore interacts with both the chromosome and a microtubule filament, which pulls or “segregates” the chromosome to one side of the cell during mitosis (Figure). In the KaryoCreate system, centromere-specific gRNA-protein complexes should disrupt the interaction between microtubules and specific chromosomes, thus preventing proper segregation.
Development Stage
The inventors are currently developing strategies to increase the frequency of chromosome mis-segregations using this technology.
Applications
- Generate cell lines with specific aneuploidies: This technology facilitates studies of specific aneuploidies in the context of human cancer or other diseases.
- Study basic chromosome segregation biology: Can use this system to study the basic mechanisms of chromosome segregation.
Advantages
- Specific chromosome gains/losses: Targeting unique centromere sequences allows for studies of specific chromosome aneuploidies. Existing techniques for studying aneuploidy are not chromosome-specific and induce random chromosome mis-segregations.
- Applicable to cells from different tissue types, cancer types, and non-tumorigenic tissues: This technology has been validated in hCEC, HCT116, LoVo, LS513, SW48, CL-11, RPE-1 cell lines.
Intellectual Property
NYU has filed for a pending PCT application covering the composition and methods of generating aneuploidy with KaryoCreate technology (PCT/US2023/073784).
Cells Lines Available for Licensing
- LoVo Colorectal Cancer: Chr 7-trisomic/ 7-disomic | Chr 5-trisomic/ 5-disomic
- LS513 Colorectal Cancer: Chr 7-trisomic/ 7-disomic
- Human Colonic Epithelial Cell (hCEC): control sgRNA | Chr 18q loss | Chr 7 gain | Chr 6 | Chr 8 | Chr 9 | Chr 12| Chr 18| Chr13 + 21| Chr X